| «Japanese» «English» | |
| October 15 (Mon) October 16 (Tue) October 17 (Wed) | |
| October 16 (Tue) | |
| ◆Big Hall | |
| <Invited Talks: Chemical Biology> 10:00-11:30 | |
| Hiroshi Handa (Tokyo Institute of Technology, Graduate School of Bioscience and Biotechnology) | |
| Title: Expansion from chemical target identification to drug discovery | |
| Abstract: Low-molecular compounds including pharmaceutical agents selectively bind to target proteins in the body and demonstrate medicinal effects by altering the function and structure of the targets. Accordingly, identification of targets is absolutely essential for understanding the mechanism of action of the drug and the network related to the target. We have developed a novel magnetic-bead technology which enables one-step isolation/identification of a target protein from various libraries which are mixtures of more than several hundreds of thousands of proteins. It is, however, not easy to develop new drugs based on the identified target. In this symposium, I would like to share our findings so that we can discuss in open debate on the potential of target identification leading to drug discovery. | |
| Minoru Ueda (Tohoku Univ.) | |
| Title: Chemical Biology of Glycoside Natural Products | |
| Abstract: Glycoside natural products have been considered as minor bioactive compounds. This is because glycosylation of small compounds always cause a severe decrease in bioactivity. It has been widely recognized that glucosides are synthesized as storage or waste forms. On the other hand, glycosylation is indispensable for the expression process of bioactive membrane proteins. Is there any possibility that we can find similar biological importance of glycosylation against natural products? This talk will present such examples in bioactive natural products. We found that jasmonate glucoside, which is a glucoside of jasmonate (an oxylipin-type phytohormone), functions in a totally different mode of action as does the original ligand. Glycosylation of JA can completely switch its target from cytosolic COI1-JAZ receptor complex to membrane protein MTJG. We named this novel mechanism for the regulation of bioactivity “glycosylation switching”, because drastic transformation of bioactivity from the original ligand into its glucoside form behaves much like the flipping of a switch. This is an unexpected and surprising role of glycosylation in the regulation of bioactivity. I will also present another possibility of “glycosylation switching” that may operate in a human body. |
|
| <Invited Talks: Perspective of in-silico drug design 1 > 13:30-15:00 | |
| Yuji Mochizuki(Rikkyo Univ.) | |
| Title: Development of ABINIT-MP(X) program for FMO calculations | |
| Abstract: The ABINIT-MP(X) program has been originally developed by our group for FMO calculations. In this talk, the features and performances of this program will be summarized as well as some illustrative examples. | |
| Hiroshi Chuman(Tokushima Univ.) | |
| Title: Development of a Novel QSAR using Molecular Calculations: Linear Expression by Representative Energy Terms | |
Abstract: 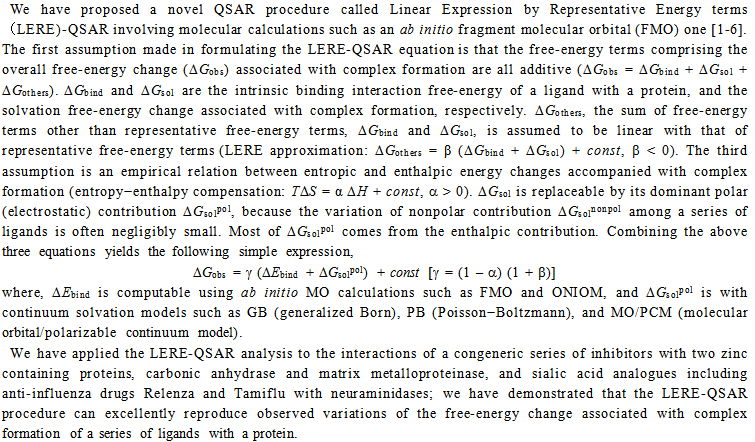 |
|
| <Invited Talks: Perspective of in-silico drug design 2 > 15:30-17:00 | |
| Osamu Ichihara (Schrödinger K.K.) | |
| Title: Understanding the role of hydration in ligand binding: Application to rational drug design | |
| Abstract: In recent years, the contribution of hydration to the thermodynamics of ligand binding has been a topic of considerable interest in drug discovery research. The computational tool WaterMap (Schrödinger LLC, New York, 2008) clusters solvent positions from molecular dynamics simulations into high occupancy hydration sites and calculates their enthalpies and entropies with respect to bulk solvent. We studied the effect of hydrating water molecules on the binding of over 20 fragment hit/lead compound pairs (CDK2, p38, HSP90, uPA, PPARs, BACE1, PDE4 etc) associated with X-ray structural data found the literature† using WaterMap. The analysis revealed that there were distinct differences between the thermodynamic profiles of the water molecules displaced by fragment hits and lead compounds. Fragment hits tend to displace waters with higher contribution of the entropy term, but not necessarily the waters with larger total free energy gain compared with the corresponding leads. Details of the analysis and the implications to fragment hit optimization strategy will be discussed. † Orita M., Ohno K., Warizaya M., Amano Y., Niimi T., Methods in Enzymology 2011, 493, 384 |
|
| Tomonaga Ozawa (Kissei Pharmaceutical Co., Ltd.) | |
| Title: | |
Abstract:  |
|
| Naoki Nakao (DAIICHI SANKYO CO., LTD.) | |
| Title: Utilization of Computer-Aided Drug Design in pharmaceutical industry | |
| Abstract: With the rapid progress of computational technology over the past decade, a concept of Computer-Aided Drug Design (CADD) become widely applied in drug discovery research and bench chemists are using CADD software for themselves. However, it does not seem to reach a sufficient level yet because of serious difficulties for precise prediction of ligand – protein interaction. I will introduce current activities of CADD in our company and discuss our efforts to overcome the issues. | |
| ◆Small Hall | |
| <Invited Talks:Utilization of K computer in Life Science: Future Visions and Issues > 13:30-15:00 | |
| Takashi Ishida (Tokyo Inst. of Tech.) | |
| Title: GHOST-MP: an ultra-fast computing pipeline for metagenome analysis on K supercomputer | |
| Abstract: Metagenome analysis is useful for not only understanding symbiotic systems but also watching environment pollutions. These days, next-generation sequencer (NGS) became to produce a huge amount of genomic data in a short time and it is expected that metagenomic researches are promoted based on such huge data. However, metagenome analysis requires sensitive sequence homology search processes because current databases do not include sequence data for most of microbes in a sample. Unfortunately, these processes need large computation time and are thus a bottleneck in current metagenome analysis. Here, we developed a large-scale computing pipeline for analyzing a huge amount of metagenomic data obtained from a NGS. The pipeline enables us to analyze metagenomic data by utilizing huge computational resources on K-computer. Also, we developed a fast homology search algorithm based on suffix array. By using these new program and pipeline, we can process metagenome data obtained from a NGS in a few hours. | |
| Shu Takagi (The Univ. of Tokyo) | |
| Title: The development of multiscale thrombosis simulator toward the predictive medicine | |
| Abstract: Thrombosis is an important circulatory disease, which causes the myocardial and cerebral infarctions. Platelet aggregation, which is the initial stage of thrombus formation, is classified into two categories, the primary and the secondary aggregations. The primary aggregation starts with the adhesion of platelet and gives the activation of platelets. The secondary aggregation follows after this activation and brings the irreversible process toward the thrombus formation. At the primary aggregation, the protein-protein interaction between glycoprotein Ibα (GPIbα) on platelet surfaces and von Willebrand Factor (vWF) on the injured vessels plays an important role. Starting from this stage, the entire process of thrombus growth is very complicated and shows a typical multiscale problem, affected from molecular scale protein-protein interaction to continuum mechanical scale in blood flow. This requires the large scale coupling method for different scales. Related to this phenomenon, we have been developing the multiscale thrombus simulation for “K” computer. The numerical model simulating the molecular scale interaction between platelets and vascular endothelium is introduced through the stochastic Monte Carlo simulations. Then, the interacting force obtained from the Monte Carlo simulation is coupled with the continuum scale blood flow simulation using finite-difference based fluid-structure-interaction method. This multiscale thrombus simulator is explained in the present talk. | |
| Takaharu Mori (RIKEN) | |
| Title: Molecular simulations of membrane proteins: Towards an understanding of their biological functions | |
| Abstract: Membrane proteins are encoded by up to 30% of genes in most organisms and are a major target for drug design. Therefore, their structures and functions have been extensively studied both experimentally and theoretically. Molecular dynamics simulation is a powerful research tool to explore how conformational dynamics impacts on the function of membrane proteins. Since the size of protein-membrane systems is generally large, it is necessary to use a massively-parallel supercomputer for the simulations. In the presentation, we show our recent studies on the molecular dynamics simulations of the protein-conducting channel (Sec translocon) 1,2). We also introduce our new simulation technique and its application to biomembrane systems. 1) T. Mori, R. Ishitani, T. Tsukazaki, O. Nureki, and Y. Sugita, Biochemistry, 49, 945-950 (2010). 2) T. Mori, F. Ogushi, and Y. Sugita, J. Comput. Chem., 33, 286-293 (2012). |
|
| Chisa Kamada (RIKEN) | |
| Title: K computer and SCLS computer system open to researchers of life sciences | |
| Abstract: The High Performance Computing Infrastructure (HPCI) encompasses the K computer and other computational facilities at nine Japanese universities. The MEXT-funded HPCI Strategic Program Computational Life Science and Application in Drug Discovery and Medical Development has promoted utilization of the K computer by life science researchers to further its “Establishment of Human Networks” objective. In early 2013, the supercomputer system with the K computer (SCLS computer system) will be made available to the life science research community. The Computational Life Science and Application in Drug Discovery and Medical Development program and its activities will be presented. In addition, the capabilities of the SCLS computer system will be described, together with the support services available to computational life science researchers and the larger biomedical research community. |
|
| <Invited Talks: Integrated omics approaches in medicine > 15:30-17:00 | |
| Norie Araki (Kumamoto Univ.) | |
| Title: Disease systems biology based on integrated proteomics for studying cancer mechanism | |
| Abstract: Advanced integrated proteomics of disease related tissue/cellular proteins have the advantage of giving direct information on their specific signals and mechanisms. We recently established the Proteomics Research Core Laboratory which utilizes a series of sensitive and high-throughput MS systems and developed sequential quantitative proteomic strategies by using LC-shotgun (iTRAQ), 2D-DIGE and combined with DNA array. To process those voluminous data arising from each analysis, we created the “iPEACH” (PCT/JP2011/58366) application to integrate information from several analysis types into a useful data file that provides comprehensive proteomic data, including post-translational modification, transcriptomic data, and functional annotations from several databases. For expediting data interpretation, a simple tool for integration of proteomic GO analysis (MANGO, MCP 2009) and a combined method with cellular signal network analysis were also constructed to readily extract and organize the proteins most likely involved in the biological process of interest. We utilized this combination system for the analysis of molecular mechanism of gliomas including their stem cells to identify their specific proteins functionally related to the tumorigenesis and chemotherapy sensitivities. Biological validations of some core proteins extracted in the activated signal group of 200 specific tumor related proteins in 8000 proteins identified in this system revealed that these proteins have novel and specific roles in the differentiation, apoptosis, metastasis, and chemoresistance. These abnormal cellular signals, activation loops, identified were expected to be candidates of chemotherapy targets. The data indicate that our sequential proteomic strategy is standardizable for targeting and elucidating the functions of proteins involved in significant cellular events. The practical advantages of these proteomic strategies and a series of our successful results studying the molecular mechanism of cancers will be introduced. | |
| Junichi Kamiie (Azabu Univ.) | |
| Title: Development of AQUA protein for simultaneous assay of precision quantification of protein expression level, identification and quantification of post-translational modification | |
| Abstract: Expression level and post-translational modification (PTM) profile of protein are fundamental information of life science. We have developed absolute quantitative assay with isotope labeled full-length proteins as references (AQUA protein). This method can quantify proteins with high accuracy by using full-length protein as references. Also, AQUA protein makes it possible to obtain PTM profile of protein by differential analysis for quantitative values of each peptide. In this lecture, I will explain the detail of AQUA protein and present our study of renal pathology with AQUA protein. | |
| Nobuhiro Zaima (Kinki Univ. Faculty of Agriculture) | |
| Title: Application of metabolomic imaging to biology, medicine, agriculture and pharmacology | |
| Abstract: Matrix-assisted laser desorption/ionization imaging mass spectrometry (MALDI-IMS) is attracting attention as a new valuable tool. MALDI-IMS is a two-dimensional MALDI mass spectrometric technique used to visualize the spatial distribution of molecules without extraction, purification, separation, or labeling of biological samples. MALDI-IMS has revealed the characteristic distribution of several biomolecules, including proteins, peptides, amino acids, lipids, and carbohydrates, in various tissues. The versatility of MALDI-IMS has opened a new frontier in several fields, such as biology, medicine, pharmacology, pathology, and agriculture. In this presentation, I would like to introduce the applications and issues of MALDI-IMS for biological samples. | |
| Kazuhiko Uchida (Faculty of Medicine, Univ. of Tsukuba) | |
| Title: Development of biomarker for preemptive medicine by OMICS analysis | |
| Abstract: We can prevent disease progression if it is diagnosed at an early stage, and as a result, if early intervention for disease is applied. The number of people with dementia is three million in Japan right now, and it is predicted it will be more than 100 million people in worldwide in 2050. Early treatment and intervention at preclinical stage of dementia should greatly reduce the number of patients. In this way, the diagnosis and intervention at preclinical status is preemptive medicine, and biomarker discovery is a key to achieve it. We have developed the biomarker discovery system consisting of multidimensional HPLC, mass spectrometry, and differential analysis software tool and we have been conducting search biomarkers for early stage of disease. Omics analyses of post-translational modifications such as glycosylation and low molecular weight proteomics revealed potential biomarkers in blood. Possibility of paradigm shift from curative to preemptive medicine by development of such biomarkers will be discussed. | |
